Libraries
| Library | Name | Clones | Raw ESTs | High quality ESTs | Mean EST length | std EST length |
| 1 | GRZ_Bank1_Sanger | 131808 | 131808 | 129826 | 698.6 | 136.9 |
| 2 | GRZ_Bank1_454 | 751885 | 751885 | 662494 | 212.2 | 42.6 |
| 3 | GRZ_Bank2_454 | 415426 | 415426 | 342570 | 218.3 | 40.7 |
| 4 | GRZ_Bank3_454 | 2351236 | 2351236 | 2136328 | 367.6 | 101.2 |
| 5 | GRZ_Bank5_Illumina | 89730910 | 89730910 | 89730910 | 151.0 | 0.0 |
| 6 | GRZ_Bank6_Illumina | 55410042 | 55410042 | 55410042 | 101.0 | 0.0 |
| 6 | GRZ_Bank6_Illumina | 33947044 | 33947044 | 33947044 | 76.0 | 0.0 |
| 7 | GRZ_Bank7_Illumina | 60997820 | 60997820 | 60997820 | 101.0 | 0.0 |
| 7 | GRZ_Bank7_Illumina | 36117265 | 36117265 | 36117265 | 76.0 | 0.0 |
| 8 | GRZ_Bank8_Illumina | 35963921 | 35963921 | 35963921 | 76.0 | 0.0 |
| 9 | GRZ_Bank9_Illumina | 27369310 | 27369310 | 27369310 | 76.0 | 0.0 |
| 10 | GRZ_Bank10_Illumina | 31727935 | 31727935 | 31727935 | 76.0 | 0.0 |
| 11 | GRZ_Bank11_Illumina | 34720289 | 34720289 | 34720289 | 76.0 | 0.0 |
| 12 | GRZ_Bank12_Illumina | 30268649 | 30268649 | 30268649 | 76.0 | 0.0 |
| 13 | GRZ_Bank13_Illumina | 31423327 | 31423327 | 31423327 | 76.0 | 0.0 |
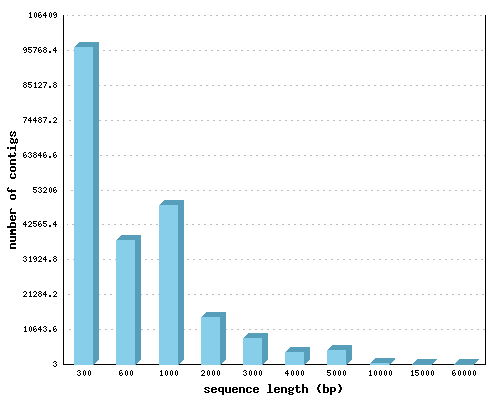
cDNA contigs
Minimum length (bp): 300
Maximum length (bp): 64116
Average length (bp): 1183.8
|  |
Annotation
Annotation statistics
Number of annotated cDNA contigs: 112853 (52.8%)
Number of full length cDNA contigs: 32813 (15.4%)
cDNA contig with blast hits
| Database | Number of contigs | % |
| any_database | 126023 | 59.0 |
| ensembl_fish_cdna | 100123 | 46.9 |
| ensembl_fish_proteins | 101309 | 47.4 |
| genage_human | 14286 | 6.7 |
| ncbi_fish_unigenes | 110267 | 51.6 |
| nr | 100369 | 47.0 |
| refseq_human_proteins | 82719 | 38.7 |
| uniprot | 101804 | 47.7 |
| Any database | 126023 | 59.0 |
cDNA contigs with predicted structural domains
| Database | Number of contigs | % |
| any_database | 38461 | 18.0 |
| pfam | 38461 | 18.0 |
| Any database | 38461 | 18.0 |
cDNA contigs with orthologues
| Database | Number of contigs | % |
| any_database | 0 | 0.0 |
SSRs and Contamination
SNP and SNV statistics
SNP transversions: 47651 (38.5%)
SNP transitions: 76123 (61.5%)
SNV transversions: 44270 (38.4%)
SNV transitions: 70948 (61.6%)
| Mutation | Number of SNP |
| A<->C | 6437 |
| A<->G | 18840 |
| A<->T | 7255 |
| C<->A | 6124 |
| C<->G | 4463 |
| C<->T | 19607 |
| G<->A | 19152 |
| G<->C | 4224 |
| G<->T | 6307 |
| T<->A | 6883 |
| T<->C | 18524 |
| T<->G | 5958 |
| Mutation | Number of SNV |
| A<->C | 5673 |
| A<->G | 17132 |
| A<->T | 6921 |
| C<->A | 6210 |
| C<->G | 3851 |
| C<->T | 19194 |
| G<->A | 18512 |
| G<->C | 3556 |
| G<->T | 6342 |
| T<->A | 6460 |
| T<->C | 16110 |
| T<->G | 5257 |
Number of transcripts with indels: 0 (0.0%)
Microsatellites detected by sputnik
| Location | Number of microsatellites |
| 3 utr | 10725 |
| in orf | 9646 |
| 5 utr | 6193 |
| other | 1315 |
| Total | 27879 |
| Unit | Number of microsatellites |
| AC | 12371 |
| A | 2258 |
| AGG | 1476 |
| AAT | 1357 |
| AG | 1246 |
| ATC | 871 |
| AT | 750 |
| AAAT | 733 |
| AAG | 642 |
| ATCC | 563 |
| AGC | 542 |
| C | 429 |
| AAC | 368 |
| AAAC | 347 |
| AGAT | 282 |
| AAAG | 262 |
| AATC | 253 |
| ACC | 227 |
| ACGC | 221 |
| AATTC | 218 |
| AAAAT | 210 |
| AATG | 199 |
| ACTC | 185 |
| ACT | 158 |
| AATAG | 142 |
| ACAT | 129 |
| ACAG | 127 |
| AAAAC | 99 |
| ACCAT | 83 |
| AAAAG | 69 |
| AATGG | 68 |
| AGGG | 67 |
| AACT | 54 |
| AAGG | 48 |
| ACG | 43 |
| AAGT | 42 |
| AAATC | 37 |
| AACC | 30 |
| AGAGG | 29 |
| AATCC | 27 |
| CCG | 24 |
| AACAC | 22 |
| AGCGG | 22 |
| AAGAT | 22 |
| AAATT | 20 |
| AACTG | 19 |
| ACGAG | 19 |
| ACCGG | 18 |
| AATAT | 17 |
| AGGC | 17 |
| AATCT | 17 |
| ACACT | 16 |
| AAACT | 16 |
| ACTG | 15 |
| ACTCT | 15 |
| AAGCT | 15 |
| AACAT | 15 |
| AAACC | 14 |
| ACCCC | 14 |
| ACCT | 13 |
| ACACC | 12 |
| AGCT | 12 |
| ACTCC | 12 |
| AATT | 12 |
| AGCCT | 10 |
| ACCCG | 10 |
| ACAGG | 9 |
| ACGT | 9 |
| ACGAT | 9 |
| AAATG | 8 |
| CG | 8 |
| ACCAG | 7 |
| AACCC | 7 |
| AACAG | 7 |
| AGATC | 7 |
| AGGGC | 7 |
| AGCTC | 6 |
| AGATG | 6 |
| ATGCC | 6 |
| AATGT | 6 |
| ACGG | 6 |
| AGCG | 6 |
| AAGAG | 5 |
| ACCTC | 5 |
| ACGCC | 5 |
| AAAGT | 5 |
| AGCAT | 5 |
| ACTAG | 5 |
| AACCT | 4 |
| AGGCG | 4 |
| AACG | 4 |
| AAGTG | 4 |
| ACCTG | 4 |
| ACATC | 4 |
| AACGT | 4 |
| ACCCT | 4 |
| ACTAT | 4 |
| AATAC | 4 |
| AACTT | 4 |
| AAGGC | 4 |
| CCCCG | 4 |
| AAGAC | 3 |
| ACCC | 3 |
| CCCGG | 3 |
| AGAGC | 3 |
| AGCCG | 3 |
| AGCC | 3 |
| AACGC | 3 |
| AACCG | 3 |
| AAGGG | 3 |
| AAGTC | 2 |
| AGGGG | 2 |
| ACAGC | 2 |
| AGCCC | 2 |
| ACAGT | 2 |
| ACCGT | 2 |
| ACGGG | 2 |
| ACGCT | 2 |
| AGGAT | 1 |
| ACATG | 1 |
| AACTC | 1 |
| AAAGG | 1 |
| ATATC | 1 |
| AGCGC | 1 |
| AGGCC | 1 |
| Total | 27926 |
Contamination statistics
| Origin | Number of contigs |
| fungi | 154 |
| invertebrate | 1495 |
| microbial | 21 |
| plant | 581 |
| protozoa | 926 |
| vertebrate_mammalian | 4208 |
| Total | 7385 |

|
Beutenbergstraße 11 D-07745 Jena • Germany |
Phone: +49 3641 65-6000 Fax: +49 3641 65-6351 |
E-mail:
info@leibniz-fli.de
www.leibniz-fli.de |
Imprint
Data Privacy |

|
